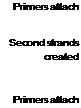 |
 |
The use of ELISA, RT-PCR, and Northern and Western blot analyses are very useful in identifying specific inflammatory mediators which are inhibited by anti-inflammatory compounds. However, when one is designing topical formulations for treating an inflammatory skin condition, it is not only necessary to identify the inflammatory mediators that can be inhibited by topical application of a lotion or gel containing a given
Second strands created
Figure 8 Diagram of the polymerase chain reaction (PCR) showing how gene sequences are amplified.
anti-inflammatory compound, but it is also important to have some knowledge of what beneficial genes and proteins may be inhibited by the topical product. For example, although corticosteroids are potent anti-inflammatory agents when used topically, they have negative side effects including the inhibition of collagen production in the skin, the reduction of the immune response to a point where a risk of skin infections increases, and at high doses, inhibition of fibroblast proliferation. Thus, to develop an effective and safe topical anti-inflammatory product that does not damage skin structure and function, it is important to determine what potentially beneficial genes in keratinocytes, fibroblasts, and immune cells, for example, IL-10, collagen III, or tissue inhibitor of metalloproteinase (TIMP), may be suppressed by the anti-inflammatory compound. One of the most effective methods for screening anti-inflammatory compounds for both their positive and negative effects on gene expression is the use of gene array technology (72). With this technique it is possible to assess the expression level of hundreds to thousands of genes simultaneously. Gene arrays are membrane filters or glass slides to which are bound small pieces of known and/or unknown (EST-expressed sequence tags) human genes. A typical nylon gene array filter may contain as few as fifty or as many as 5000 different gene sequences on a single filter, and some arrays have even been designed with specific tissues or diseases in mind, such as inflammation. The sequence of steps involved in a gene array analysis is shown in Figure 9. The first step involves isolating mRNA from untreated cells (control group) and from cells exposed to some experimental condition (experimental group). After hybridization, any unbound cDNA is washed away and the hybridized cDNA is detected and quantified. Since the location and identity of each gene on the filter is known, by comparing the quantified spots on the array produced from the control group to those spots produced from the experimental group, one can determine if a particular gene in the experimental group is up-regulated or down-regulated compared to the control
 Control Experimental
Control Experimental
![]() Cells or tissue
Cells or tissue
Isolate mRNA
![]() Experimental Array
Experimental Array
Figure 9 Steps involved in gene array analysis.
group. Given the complexity of gene arrays, a computer software program is used to aid in the quantification and analysis of the large amount of data that is obtained. The software produces an “overlay” image of the filters from both the control and experimental groups, calculates the difference in expression level for each gene between the two groups, and then converts this relative expression data into a color image. For example, a gene that is up-regulated in the experimental group compared to the control group is shown as a green spot on the computer generated image, while a gene that is down-regulated in the experimental group is shown as a red spot. By using this method the effect of any compound on the expression of genes that code for pro – and anti-inflammatory mediators as well as other genes expressed in epidermal and dermal cells can be rapidly determined. An example of the use of this technology is shown in Figure 10. In this experiment human fibroblasts were treated with quercetin and the effect of this compound on the expression of inflammatory and dermal matrix altering genes determined. The cDNA array images from untreated (A) or quercetin (B) treated fibroblasts captured by the phosphoimager were merged and colorized by the computer software to yield the image in panel C. Genes that were down-regulated in quercetin treated fibroblasts relative to those in untreated cells are displayed by the software as either red or yellow while those genes up-regulated by quercetin are displayed as green. In this study, quercetin was found to lower the expression of MCP-1 and collagenase (MMP-1) while up-regulating a gene that blocks MMP activity (TIMP-1). Table 2 shows the results of an array analysis of genes that are up- and down – regulated by quercetin in TPA-treated keratinocytes. Note that quercetin up-regulates genes that are play a role in protecting the dermal matrix and down-regulates matrix destroying genes.
